News
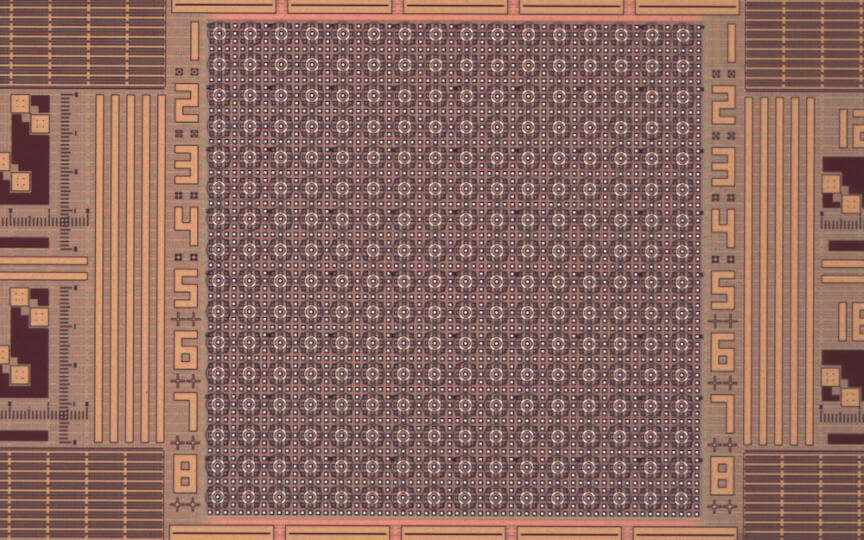
This semiconductor chip, features a dense array of microsites that can control the pH at the local level. (Credit: Donhee Ham Research Group/Harvard SEAS)
pH — the concentration of protons in a watery solution — indicates how acidic the solution is. It regulates a broad range of natural and engineered chemical processes, including the synthesis of designed DNA sequences for applications in biotechnology.
Changing the pH uniformly across an entire water-based solution is a standard practice in chemistry. But what if researchers could create an array of localized pH regions where protons are more intensely concentrated than in other parts of the solution? This would allow them to perform pH-regulated chemistry at each of those locations in parallel, dramatically increasing the experimental throughput and speeding up processes in DNA synthesis, which has applications in genomics, synthetic biology, vaccine development and other therapies, and data storage.
But localizing pH is a challenge because protons spread out fast in a water-based solution.
Now, researchers from the Harvard John A. Paulson School of Engineering and Applied Sciences (SEAS), in collaboration with researchers at the Broad Institute of MIT and Harvard, and DNA Script, a biotech focused on enabling bench-top enzymatic DNA synthesis, have developed a technique to control pH at the local level, creating a dense array of microsites where the amount of protons is 100 to 1000 times higher than the average in the rest of the solution.
“This work enables a high-throughput application of a broad range of pH-regulated chemistry, including biomolecular synthesis,” said Donhee Ham, the Gordon McKay Professor of Electrical Engineering and Applied Physics at SEAS and co-senior author of the paper.
“It was made possible by an array of micrometer-scale electrochemical cells of unique geometry fabricated on, and operated by, a semiconductor integrated circuit chip,” said Hongkun Park, the Mark Hyman Jr. Professor of Chemistry and Professor of Physics and co-senior author of the paper.
The research is published in Science Advances.
The semiconductor chip, featuring 256 electrochemical cells on its surface, is directly interfaced with a water-based solution of quinone molecules. Each cell looks like a bullseye with two concentric metallic rings. The interior ring injects a current into the solution to electrochemically produce protons from quinone molecules. These locally generated protons attempt to spread out but are neutralized near the exterior ring that electrochemically produces base molecules from quinone molecules by pulling a current from the solution. The locally generated protons thus are trapped in and around the center of the bullseye, creating an acidic microenvironment with a lowered pH.
“Essentially, in each activated electrochemical cell, we set up an electrochemical wall using the outer ring, which the acid generated by the inner ring cannot penetrate,” said Han Sae Jung, a graduate student at SEAS and co-first author of the paper. “Since each cell is controlled independently by the underlying semiconductor chip, we can lower pH at any arbitrary subset of the 256 electrochemical cells we choose to activate. The unique cell structure we have developed on the semiconductor electronic chip enables this spatio-selective pH programming.”
“Our device can not only localize and precisely tune pH by adjusting the currents of the concentric rings of each electrochemical cell but can also monitor pH in real time using on-chip pH sensors distributed across the electrochemical cell array,” said Woo-Bin Jung, a postdoctoral fellow at SEAS and co-first author of the paper. “Therefore, we can create any spatial pattern of target pH values, or pH topography, in the aqueous solution, with the real-time feedback from the map of the spatial pH pattern we image.”
“While traditional chemical DNA synthesis is done in non-aqueous media, enzymatic DNA synthesis in aqueous media is rapidly gaining interest, as it minimizes molecular damage and hazardous waste generation and can increase synthesis speed and performance,” said Xavier Godron, the CTO of DNA Script and a co-author of the paper. “Our manipulation of spatial patterns of pH in aqueous media thus can lead to high-throughput enzymatic DNA synthesis, with many biotechnology applications from protein engineering and antibody screening to DNA information storage."
“This work shows the power of multi-disciplinary approaches bringing together semiconductor electronics, electrochemistry, and molecular biology. The technology paves the way for a range of additional biological applications including oligo libraries for diagnostics and synthetic biology-based enzyme development,” said Robert Nicol, Senior Director of Technology Development at the Broad Institute and a co-author of the paper. “Integrating these diverse disciplines required highly collaborative teams willing to learn from each other across industry and academia."
Other co-authors of the research include Jun Wang, Jeffrey Abbott, Adrian Horgan, Maxime Fournier, Henry Hinton, and Young-Ha Hwang. The research was supported by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), under contract 2019-19081900002.
Topics: Applied Physics, Bioengineering, Electrical Engineering
Cutting-edge science delivered direct to your inbox.
Join the Harvard SEAS mailing list.
Scientist Profiles
Donhee Ham
John A. and Elizabeth S. Armstrong Professor of Engineering and Applied Sciences
Press Contact
Leah Burrows | 617-496-1351 | lburrows@seas.harvard.edu