News
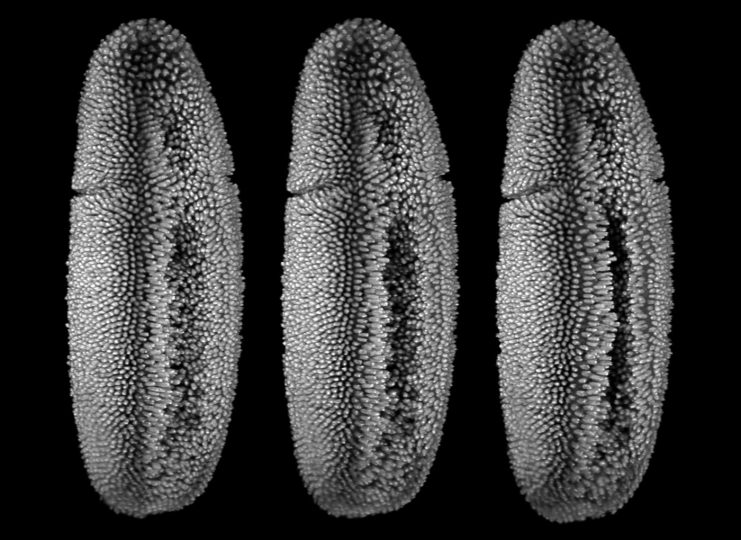
Snapshots from a live recording of Drosophila embryonic development. (Image courtesy of Stefan Günther/EMBL Heidelberg)
Embryogenesis — how an organism arises from a single cell — is one of the most mysterious and complex processes in nature. The large-scale, coordinated and collective movements of cells in a tissue during embryogenesis resemble the complex and chaotic flows of fluids in the ocean or atmosphere. But how do these movements determine which cells are destined to become part of the brain, the gut or the limb? If we could predict the fate of cells, we might be able to spot pathologies in the earliest stages of development.
Now, scientists at the Harvard John A. Paulson School of Engineering and Applied Sciences (SEAS) have applied techniques of fluid dynamics and chaos theory to embryogenesis. With this approach, they developed a framework to quantify the fate and dynamic organization of cells into tissues from imaging data.
The research was published in the Proceedings of the National Academy of Sciences.
“Understanding how geometry arises from genetics, meaning how the body makes itself using molecular signals, is one of the great questions of biology,” said L Mahadevan, de Valpine Professor of Applied Mathematics, Physics and Organismic and Evolutionary Biology, and senior author of the paper. “Recognizing the resemblance between cell movements that form tissues and the complex flows of fluids that lead to large-scale coherent motions, we devised a mathematical framework to uncover the dynamic organizing structures associated with morphogenesis.”
“This research improves our understanding of embryogenesis because instead of looking at the problem from a classical, biological sense we used particular theoretical tools that haven’t been used in this field,” said Mattia Serra, a postdoctoral fellow at SEAS and first author of the paper. “By just using chaos theory and experimental cell trajectories, we can capture the early footprint of known morphogenetic features, reveal new ones, and quantitatively distinguish different phenotypes.”
Serra and Mahadevan wanted to know if they could predict the outcome of tissue morphogenesis and organ development using information from cell-motion data, just as one can predict the locations of flotsam on the surface of the ocean from fluid motions.
“One of the challenges in the field today is that there is so much data, it’s hard to know where to look,” said Serra. “With today’s microscopes, we can observe single cells, but it’s been a challenge in the field is to predict developmental outcomes of tissue morphogenesis using single cellular trajectories. What chaos theory says is that instead of chasing the complex trajectory of every single cell, you want to understand the organizers of the movement. You want a global view, rather than a local view.”
Serra and Mahadevan teamed up with two experimental groups, one led by Sebastian Streichan at the University of California at Santa Barbara which studies fruit fly embryos, and the other led by Cornelius Weijer at the University of Dundee, which studies chicken embryos.
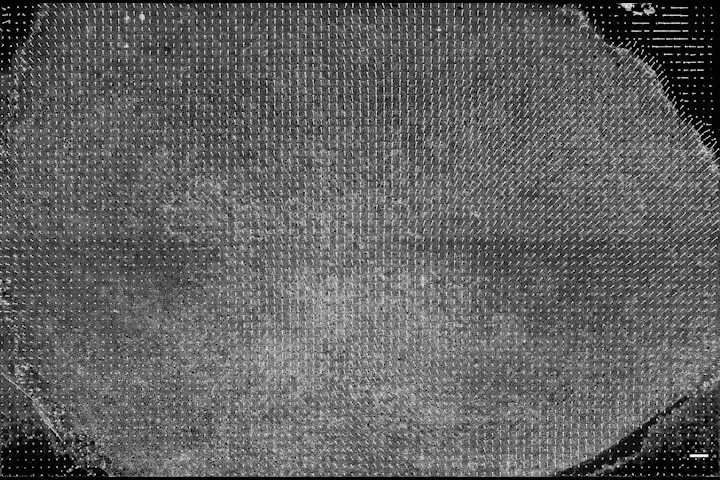
The Harvard team deployed their mathematical framework on imaging data collected by these two groups and generated a map of the embryo in space and time. This map revealed so-called spatial attractors and repellers — areas of the embryo which either attract or repel cells. These regions of attraction and repulsion appear at specific times and in specific locations during the development process.
Cell movements in a chick embryo during the formation of the primitive streak, a structure that divides a developing embryo and serves to organize embryogenesis. In a chick embryo, the formation of the primitive streak involves coordinated flow of more than 100,000 cells in the epiblast. (Video courtesy of Cornelius Weijer/Harvard SEAS)
By being able to pinpoint the exact time and location of attractors and repellers and tie them to specific morphogenetic structures like a spinal cord, the researchers could not only observe the differentiation of cells in real time, but identify the precursor cells before they began the process.
“This framework is a tool that tells you where and when to look,” said Serra. “It’s a lens that finds sweet spots where biologists can zoom in to uncover the underlying relevant mechanisms at play.”
“Our work sets the geometric stage for uncovering the dynamic organizers of cellular movements and tissue form. When combined with the ability to also track and manipulate gene expression levels, mechanical forces, etc., we will be in a position to determine the biophysical mechanisms underlying normal and pathological morphogenesis, and a little closer to answering to one of the grand questions of biology,” said Mahadevan.
This research was co-authored by Streichan, Weijer and Manli Chuai . It was supported in part by the Schmidt Science Fellowship and grants from the US National Institutes of Health
Topics: Applied Mathematics
Cutting-edge science delivered direct to your inbox.
Join the Harvard SEAS mailing list.
Scientist Profiles
L Mahadevan
Lola England de Valpine Professor of Applied Mathematics, of Organismic and Evolutionary Biology, and of Physics
Press Contact
Leah Burrows | 617-496-1351 | lburrows@seas.harvard.edu